Heegaard Floer Knot Homology
In 2007, Jean-Marie Droz of the University of Zurich (working along with Anna Beliakova) wrote a Python program to compute the (hat-version) Heegaard-Floer Knot Homology of a knot . His program is integrated into KnotTheory`, though to run it, you must have Python as well as the Python library Psycho installed on your system.
(For In[1] see Setup)
|
| ||||||||
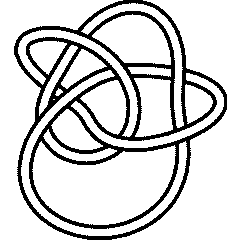
The Heegaard-Floer Knot Homology is a categorification of the Alexander polynomial. Let us test that for the knot 8_19:
In[3]:=
|
hfk = HFKHat[K = Knot[8, 19]][t, m]
|
Out[3]=
|
2 -3 m 5 2 6 3
m + t + -- + m t + m t
2
t
|
In[4]:=
|
{hfk /. m -> -1, Alexander[K][t]}
|
Out[4]=
|
-3 -2 2 3 -3 -2 2 3
{1 + t - t - t + t , 1 + t - t - t + t }
|
The knot 8_19 is the first knot in the Rolfsen Knot Table whose Heegaard-Floer Knot Homology is not "diagonal". Let us test that. The homology is "on diagonal", iff its Poincare polynomial, evaluated at , is a monomial:
In[5]:=
|
Select[AllKnots[{3, 8}], (Head[HFKHat[#][t, 1/t]] == Plus) &]
|
Out[5]=
|
{Knot[8, 19]}
|
In[6]:=
|
hfk /. m -> 1/t
|
Out[6]=
|
4 -2
-- + t
3
t
|
 K11n34 |
 K11n42 |
The Conway knot K11n34 and the Kinoshita-Terasaka knot K11n42 are a mutant pair, and are notoriously difficult to tell apart. Let us check that an array of standard knot polynomials fails to separate them, yet succeeds:
Indeed,